ZHAO Research Laboratory
Department of Chemistry
& Environmental Toxicology Graduate Program
University of California, Riverside
ZHAO Research Laboratory
Department of Chemistry
& Environmental Toxicology Graduate Program
University of California, Riverside
Copyright 2013-2026 Zhao Lab | Contact: linlin.zhao@ucr.edu
DNA damage is safeguarded by an intricate DNA repair network and several DNA damage tolerance mechanisms; nonetheless, certain DNA lesions can escape repair or exceed the repair capacity and eventually accumulate in the genome. The persisted DNA lesions can interfere with DNA transactions, leading to cell cycle arrest, mutations, and cell death. An altered genetic content in key regions, such as proto-oncogenes and tumor suppressor genes, is a major driving force for human diseases including cancer. Translesion synthesis (TLS) is a conversed DNA damage tolerance mechanism, whereby specialized DNA polymerases participate in copying past obstructive DNA structures, such as DNA lesions or non-canonical DNA structures. Because TLS is error-prone, it plays an important role in mutagenesis.




We are grateful to our sponsors


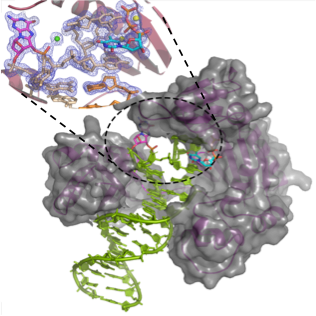
The project seeks to obtain mechanistic insights into the DNA-binding proteins and enzymes, including DNA polymerases, DNA glycosylases, and nucleases. We use enzyme kinetics, kinetic simulations, and X-ray crystallography, and mass spectrometry-based methods to dissect the basis of faulty DNA replication past DNA lesions or how DNA enzymes operate on lesion-containing DNA substrates.
Zhao et al. DNA Repair, 2024, 137, 103666.
Zhao et al. ACS Chem. Biol. 2023, 18, 5, 1168–1179.
Urrutia et al. J Biol. Chem. 2022, 289, 102306.
Xu et al. Biochemistry, 2015, 54, 639-651.
Xu et al, J Mol. Biol. 2019, 431, 673-686.
Mitochondria are vital organelles, essential for energy production, cell signaling, and protein cofactor biosynthesis in higher eukaryotic cells. Their function is critically dependent on the mitochondrial DNA (mtDNA) genome, which encodes core subunits of the electron transport chain, as well as ribosomal and transfer RNAs. mtDNA degradation is a fundamental mechanism required for mitochondrial genomic maintenance and damage response.
enzymology of DNA replication, repair, and degradation
Mitochondrial DNA degradation
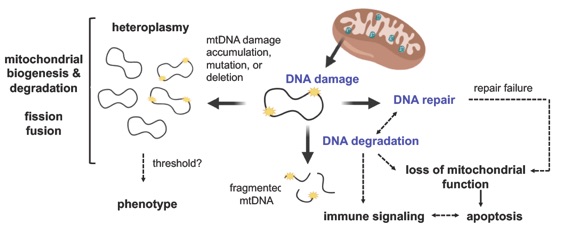
This project aims to decipher the chemical and molecular basis of mtDNA turnover. Mitochondrial transcription factor A (TFAM) is a highly abundant DNA packaging protein known for its roles in transcription and mtDNA maintenance. My laboratory is the first to demonstrate a novel role for TFAM in facilitating the degradation of damaged DNA containing abasic (AP) sites. Specifically, we found that TFAM accelerates the rate of strand breakage at AP sites by 2 to 3 orders of magnitude compared to naked DNA. Given TFAM's high abundance in mitochondrial nucleoids, this activity suggests it effectively competes with AP endonucleases in initiating damage processing. Our cellular studies confirmed this role by detecting TFAM-DNA cross-links under limiting glutathione conditions. Our working model proposes that TFAM facilitates mtDNA turnover through collaborations with nucleases and other factors. We are currently exploring biological triggers for this process, including TFAM-DNA cross-links and glutathionylated DNA adducts, and investigating their potential roles in mtDNA degradation and immune signaling.
Chen et al. PNAS, 2025
Urrutia et al. Nucleic Acids Res, 2024, 52 (22), 14093-14111.
Xu et al. Nucleic Acids Res, 2023, 1, 41-53
Zhao et al. ACS Chem Biol, 2023


Chemical Probes for sequencing DNA modifications
Recent sequencing efforts have demonstrated that the distribution of DNA modifications across the genome is highly heterogeneous. Therefore, genome-wide mapping of these modifications is critical for elucidating their roles in genetic regulation, development, and pathogenesis. The project develops DNA sequencing methodologies for mapping multiple DNA modifications with high specificity. Our innovative approach leverages the chemistry of DNA repair intermediates and develop highly specific chemical probes to achieve this goal. A recent project developed two chemicals to capture and enrich abasic (AP) sites—a central intermediate in DNA repair—and incorporate unique locator codes during amplification for high-fidelity sequencing readout. We successfully mapped both AP sites and alkylation DNA lesions within the mitochondrial DNA (mtDNA) of cultured human cells. In the long term, this high-throughput, cost-effective technology will enable the generation of single-nucleotide resolution genomic maps for multiple DNA modifications. These detailed modification maps will be invaluable for correlating specific chemical exposures or pathological conditions with distinct genomic mutation signatures.
Liu et al. Nucleic Acids Res, 2023, 51, e73
Xu et al. PNAS. 2019, 116, 17792-17799
Tang et al. Anal Chem. 2021, 93, 13398-13406
Our research program operates under two complementary themes. The first focuses on mechanistic studies of DNA enzymes and DNA/RNA-binding proteins, aiming to generate fundamental biological insights into critical cellular processes. The second theme is dedicated to translational applications, specifically developing novel chemical and enzymatic approaches to manipulate DNA and RNA processing within mitochondria for disease intervention and therapeutic use.
Chemical Probes for manipulating mitochodnrial DNA
Despite the critical role of mtDNA in human pathology and disease etiology, effective chemical strategies to prevent mtDNA loss under genotoxic stress are scarce. To address this need, we developed a mitochondria-targeting probe (mTAP) designed to selectively react with a key mtDNA repair intermediate: abasic (AP) sites. We confirmed that mTAP forms oxime conjugates exclusively with mitochondrial AP sites, showing no reaction with nuclear AP sites. Mechanistically, the mTAP conjugation rendered DNA substrates containing AP sites resistant to cleavage by AP endonuclease (APE1) and mitochondrial extracts. This conjugation significantly reduced the DNA-binding affinity of APE1 without affecting the activity of the crucial mtDNA-packaging factor, mitochondrial transcription factor A (TFAM). Crucially, cellular experiments demonstrated that mTAP treatment alleviated the decrease in mtDNA and transcription product levels induced by mitochondrial AP site damage. Furthermore, functional assays confirmed that mTAP treatment did not compromise mtDNA replication activity or increase the overall mtDNA damage level.
These findings highlight mTAP's potential as a valuable chemical tool to selectively modulate and preserve mtDNA levels under conditions of genotoxic stress.
Jana et al. Angew. Chem. Int. Ed., 2025, e202502470